The notes seldom followed a particular pattern and were often free text. Hence, In order to build this solution we first classified the notes by sectional information present in them, followed by summarizing all the clinical concepts within that note.
For summarization, we purpose-built a prompt and used it with the PaLM 2 models, such that the output is a distilled summary only containing the clinical concepts present in a discharge note.
At this point, we passed the clinical summary as an input to the PaLM2 embedding model that returns a 768-dimensional vector, which is then stored in our vector database of choice. For this solution we went ahead with the
Vertex AI Vector Search Database to create and store the vector indices.
This process may be a one time batch process or an online process as and when additional clinical data is procured.
At runtime, the solution parses the clinician's query; the query generally includes a clinical "Report" and a "Question" field, after which the parsed output is embedded using the same embedding model, i.e., the PaLM2 embedding model.
The output vector of this model is then compared against the vectors stored in the aforementioned vector database using the approximate nearest neighbor algorithm, which is built-in in Vertex AI Vector Search.
The closest neighbor vectors are then fetched and we perform a look-up to identify the clinical summaries pertaining to those closest neighbors.
These summaries are then passed as an additional input to the MedLM model, along with the existing clinical "Report" and the user's "Question," thereby enriching the query with additional contextual data.
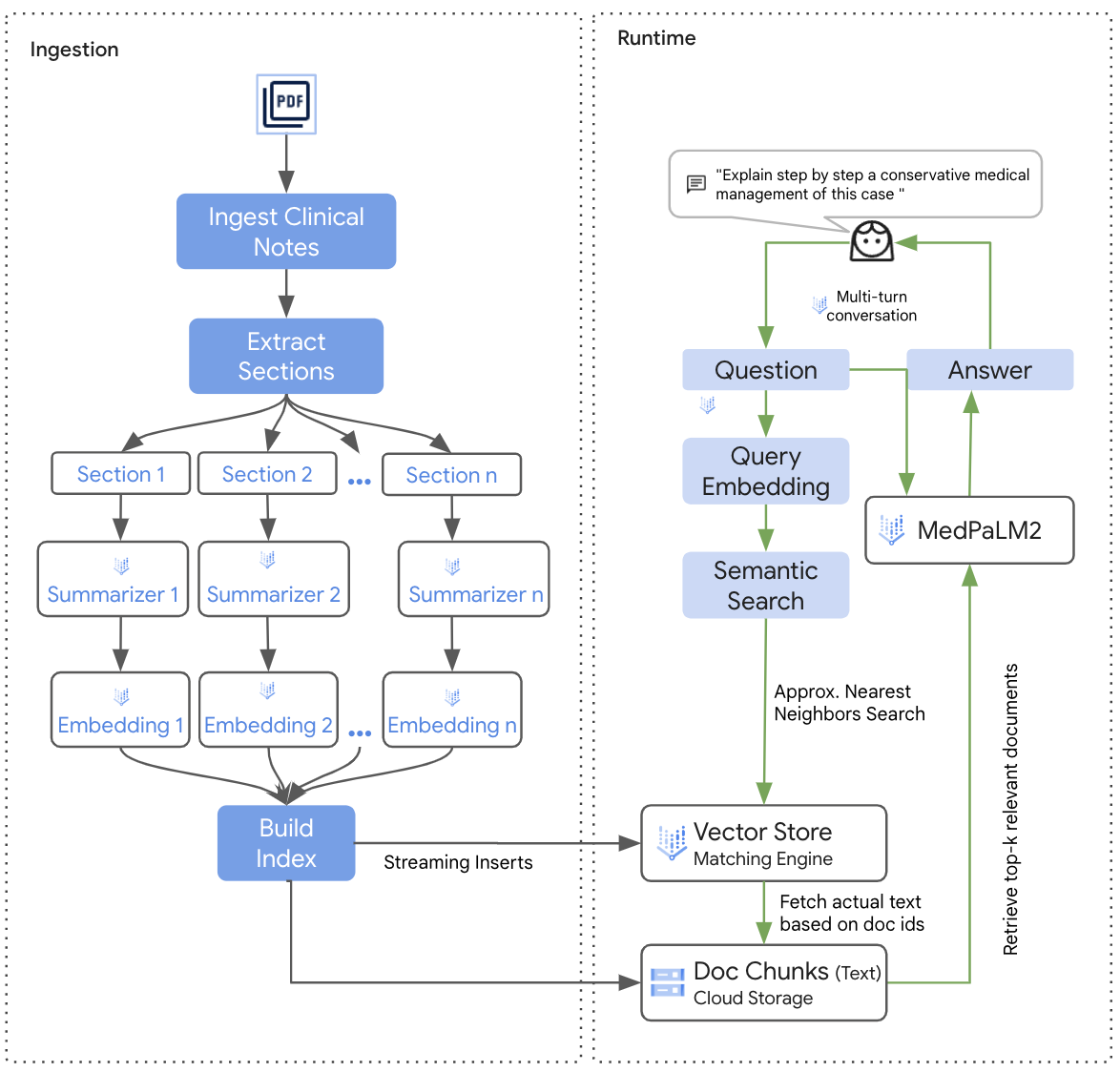 Architecture Diagram
Architecture Diagram
 Sample Result
Sample Result